Software
NovaISM NEW
Image Scanning Microscopy Analysis Software
- Seamless integration with Luminosa microscope, optimized for the PDA-23 SPAD array detector
- Higher resolution than confocal imaging with pixel-reassignment and one-click deconvolution
- State-of-the-art computational sectioning in combination with lifetime species separation, improving contrast and resolution
- Comprehensive data export options: Supports multiple formats, including OME-TIFF and scalable vector graphics
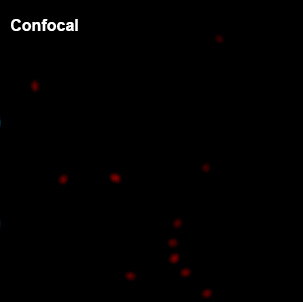
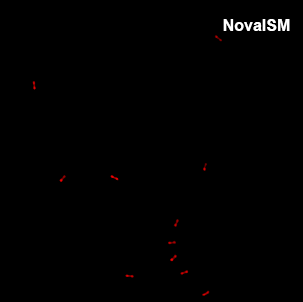
NovaISM is a powerful Fluorescence Lifetime Imaging Microscopy (FLIM) analysis software specifically optimized for the PDA-23 SPAD array detector. It combines cutting-edge pixel reassignment, computational sectioning, and deconvolution techniques to achieve up to 1.7 times higher resolution compared to conventional confocal microscopy images.
By effectively rejecting out-of-focus light, NovaISM enhances the signal-to-noise ratio and significantly improves the lifetime contrast of FLIM images. These advantages enable faster image acquisition or gentler imaging of live samples, making it an essential tool for advanced image scanning microscopy.
Image: GattaQuant 160 nm rulers (GATTA-SIM series)
Advanced ISM-FLIM analysis: High-resolution imaging with deconvolution
Quality and precision you can trust
- Computational sectioning algorithm in combination with lifetime-species separation
- Transparency over the parameters used for deconvolution and computational sectioning
Save time and simply focus on your samples
- One-Click Deconvolution
- Minimal user interaction required
- Efficient analysis of series measurements, including batch analysis and parameter plots of multiple-ROIs
Advanced flexibility
- Raw data include individual time tags of each of the 23 pixels of the PDA-23 SPAD array
- Many data export options for graphs and images (BMP, PNG, OME-TIFFf, Scalable Vector Graphics)
Images with more resolution and higher contrast
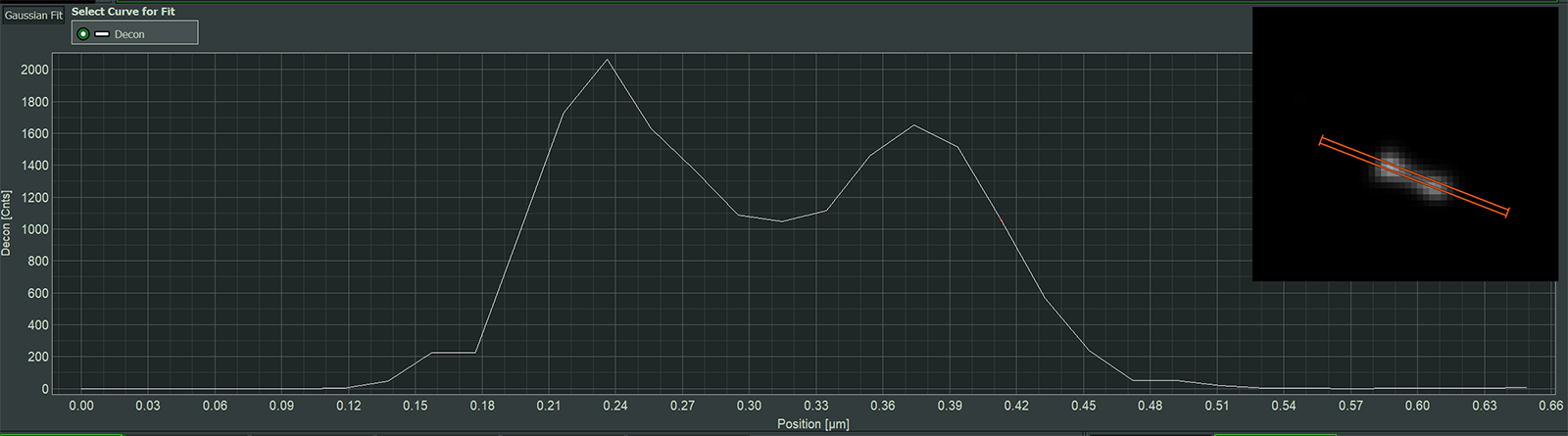
Follow the effects of ISM, computational sectioning, and deconvolution step-by-step
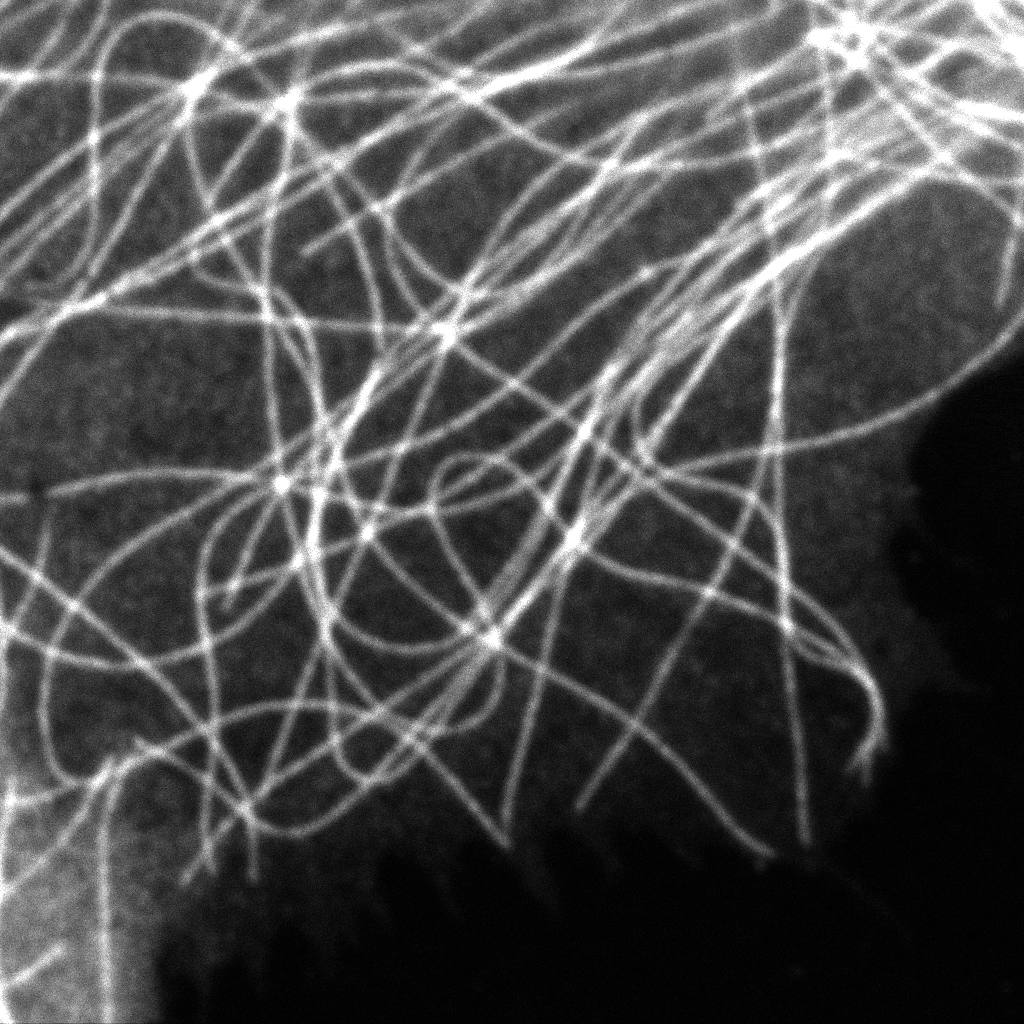
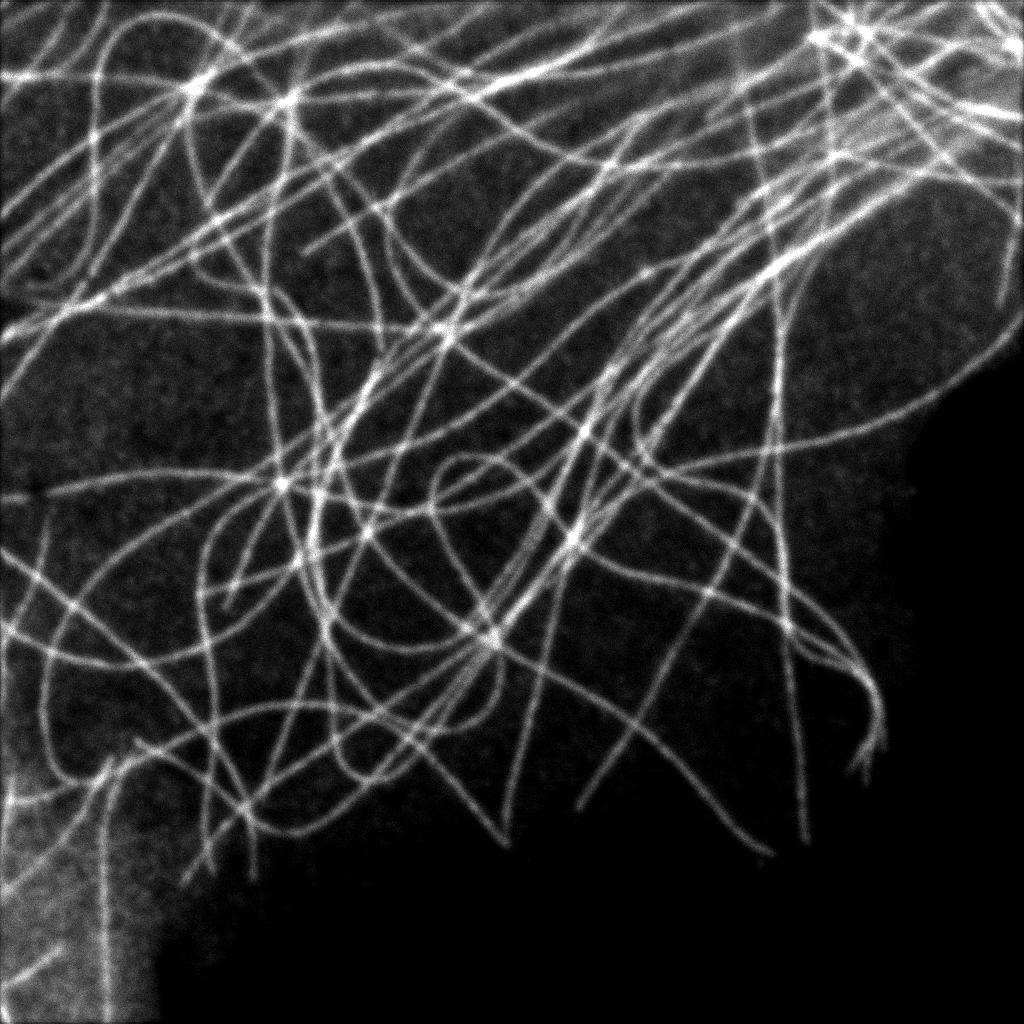
Intensity image from summing up all 23 pixels of PDA-23 Detector. This corresponds quite well to the usual confocal imaging.
U2OS Cells by GattaQuant, Tubulin staining with Alexa488.
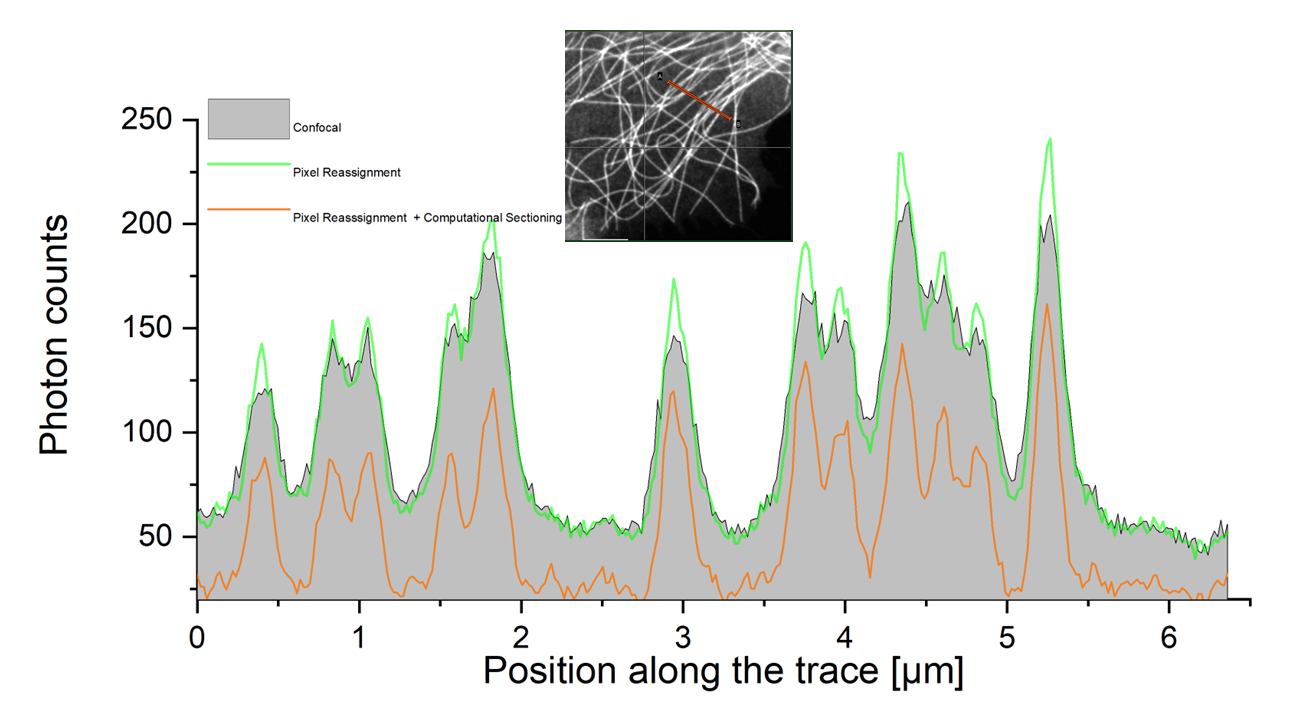
Enhanced analysis: Step-by-step improvement in intensity traces
Intensity trace along a section extending over a number of tubulin strands. The improvement achieved in each analysis step are shown.
Advanced computational sectioning: Remove out-of-focus light and separate lifetime species
The computational sectioning is based on an unmixing of the intensity distribution over the PDA-23 detector array. This process allows for rejection of light coming from out-of-focus planes which in thicker samples can be problematic as it limits the contrast, produces artifacts in the deconvolution process and interferes in the lifetime –species determination. With computational sectioning offered by NovaISM all these problems are resolved.
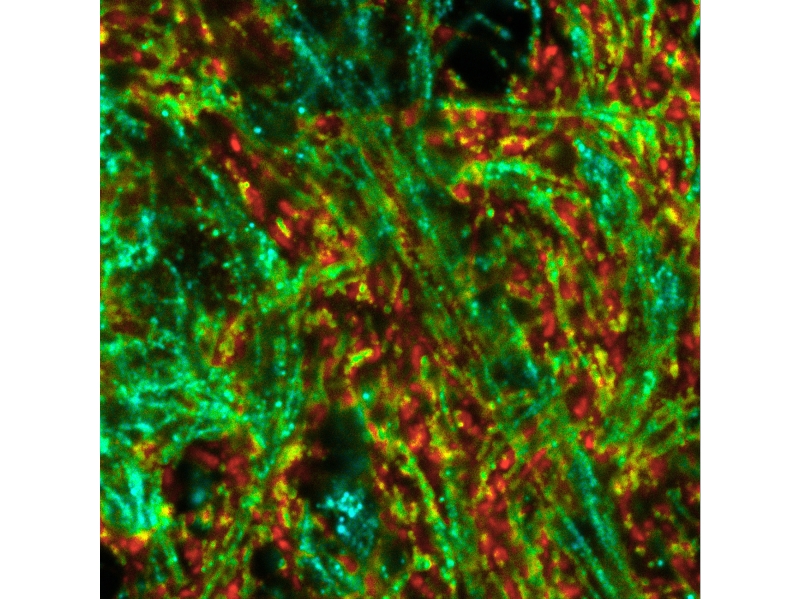
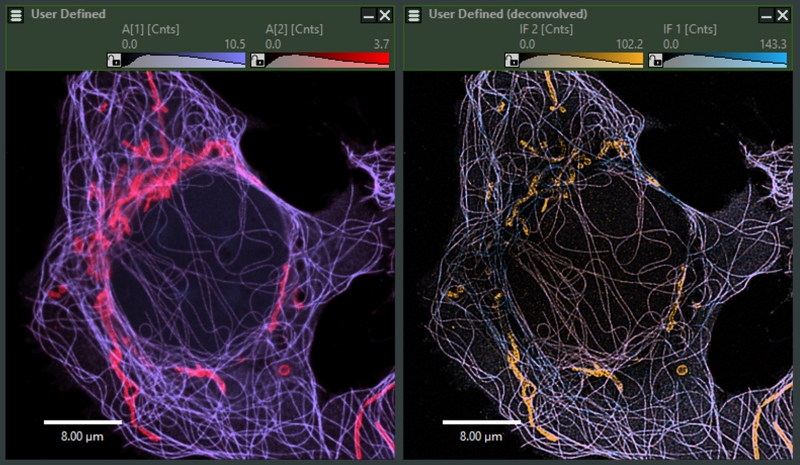
Intensity-weighted ISM-FLIM image. Colorscale indicates the fast lifetime contrast ranging from blue (shorter lifetimes) to red (longer lifetimes) sample: neuronal hippocampal culture, labeling of mitochondrial marker TOM20 with Cy2 (lifetime 1.3 ns) and synamptic vesicles marker SYPT1 with OregonGreen (lifetime 3.5 ns). Samples prepared by Dr. Eman Abbas, Rizzoli lab, UMG Goettingen.
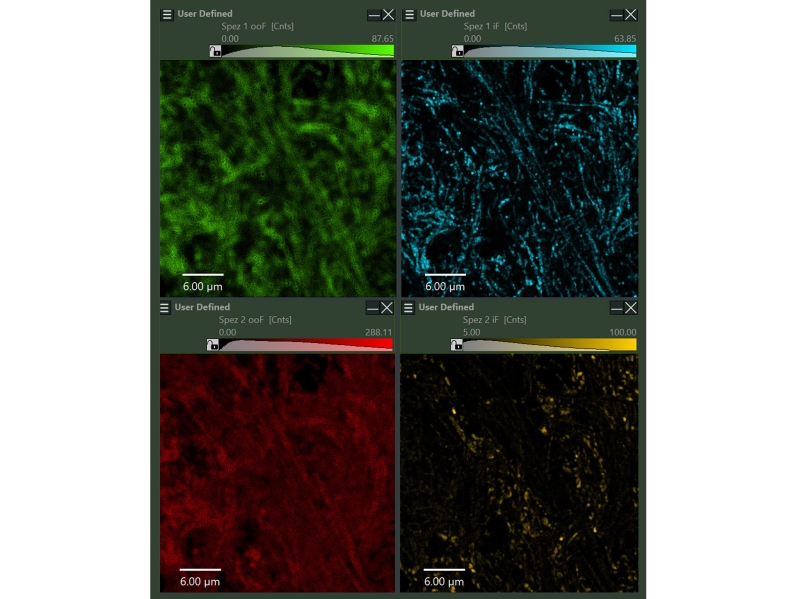
In NovaISM computational sectioning is performed simultaneously with lifetime species separation allowing for determineing in –focus and out-of-focus contributions independently for each lifetime species. For further analysis then only the infocus part can be used. Upper side: SYPT1, lower side: TOM20, left side: out-of-focus, right side: in-focus.
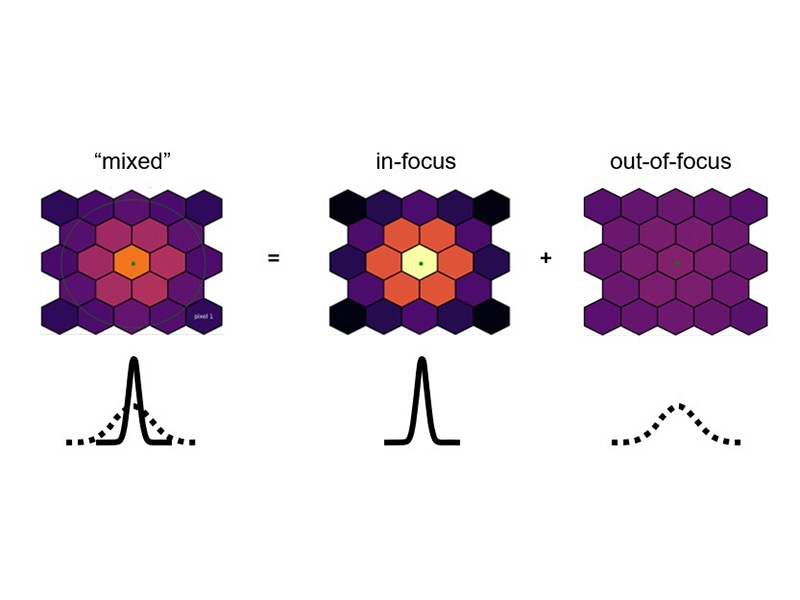
A simple schematic of the principle of computational sectioning. The distribution of light on the PDA-23 detector is fitted with 2 Gaussian of different widths. The in-focus part correspond to the narrow Gaussian and the out-of-focus part to the broader Gaussian. Parameters are estimated automatically by the software while the user can always fine tune if needed.
Request a demo of NovaISM
Contact us to make an appointment >
| Licence type | License available for either 1, 2, or 4 years | |
| Data formats | Available for *.spqr files and *.ptu files acquired with the PDA-23 SPAD Array Detector in Luminosa microscope | |
| PC system requirements | ||
|---|---|---|
| Minimal | Recommended | |
| Operating system | Win10 64-bit | Win10 / Win11 64-bit |
| CPU | Intel Core i3-8100 or AMD Ryzen 3 1300X | Intel Core i7-13700 or AMD Ryzen 7 5700X |
| RAM | 8 GB | 32 GB |
| GPU | AMD Radeon RX550/550 Series or Nvidia GeForce GT 1030 | AMD Radeon RX 6700 XT or Nvidia GeForce RTX 3060 |
| Screen resolution | QHD (2560 × 1440) | 4K (3840 × 2160) |
All Information given here is reliable to our best knowledge. However, no responsibility is assumed for possible inaccuracies or omissions. Specifications and external appearances are subject to change without notice.
| Luminosa | NovaFLIM | NovaISM | NovaFLIM+NovaISM | |
InstaFLIM
|
 |
 |
 |
 |
| Phasor plot for marker multiplexing |  |
 |
||
| Multi-frame analysis |  |
 |
||
| Batch analysis of series measurements (time-lapse,z-stacks, tiling, …) |  |
 |
||
| Advanced ROI handling, multi-ROIs, import of external ROIs |  |
 |
||
| Parameter plots of fitted parameters over multiple ROIs |  |
 |
||
| Representation of the image information as user defined 1D and 2D histograms of fitted parameters |
 |
 |
||
| ROI selection via parameter histograms |  |
 |
||
| User configured analyses allowing for FLIM, FLIM-FRET, steady-state FRET and anisotropy imaging |
 |
 |
||
| Extended export options of graphs and images |  |
 |
||
| Image Scanning Microscopy-FLIM (Resolution enhancement via a pixel reassigned image available in combination with PDA-23 Add-on) |
 |
 |
 |
|
| Re-assignement vectors calculation from image |  |
 |
 |
|
| Resolution enhancement Deconvolution for ISM-FLIM (enhanced FLIM contrast via the rejection of contribution from out-of.plane light; Available in combi with PDA-23 Add-on) |  |
 |
||
| Computational Sectioning in combination with lifetime-species seperation (enhanced FLIM contrast via the rejection of contribution from out-of.plane light; Available in combi with PDA-23 Add-on) |  |
 |

 Contact us
Contact us