Product Studies
Molecular Counting
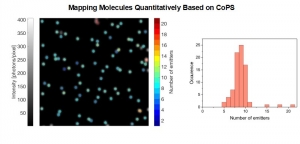
Mapping Molecules Quantitatively Based on CoPS (Counting by Photon Statistics)
- Statistical analysis of detected photon coincidences to estimate the number of independent fluorescent labels in the observation volume of a confocal microscope
- Counting and mapping of molecules and their concentrations
- Generates reliable data in the red spectral wavelength range, counting of up to 10 emitters
Description
Molecular Counting is a technique that enables quantitative mapping of the distribution of molecules using fluorescence microscopy. The approach is based on Counting by Photon Statistics (CoPS)1, which involves a statistical analysis of detected photon coincidences to estimate the number of independent fluorescent emitters in the observation volume.
Mapping molecules quantitatively based on CoPS can be carried out using a MicroTime 200 time-resolved confocal microscope equipped with four SPAD detectors and a four channel HydraHarp 400. The application also requires additional optical elements and proprietary analysis software provided by Haisen Ta / PicoQuant.
The counting method as presented here currently works best for bright and stable red emitting fluorophores in fixed samples with low background emission.
Proof of principle experiment
I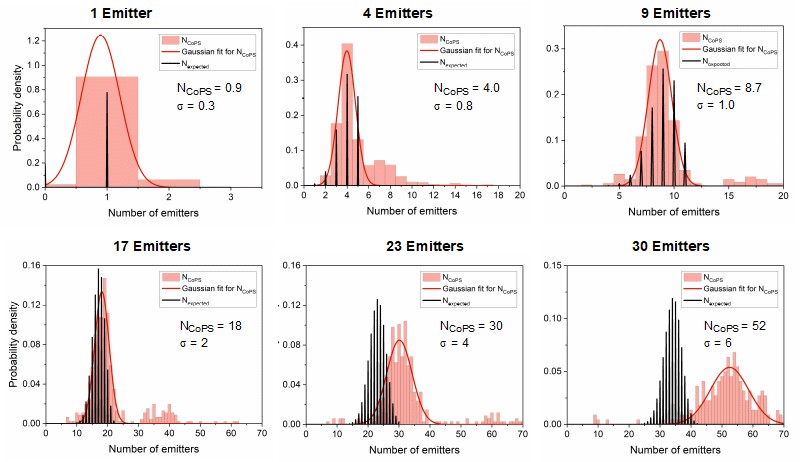 n proof-of-concept studies, DNA origamis with varying emitter numbers (from 1 to 30) per origami were quantified. The plots below show the results obtained with DNA origami emitting in the red spectral range. The black bars indicate the expected distribution pattern of emitter number per origami (assuming a binomial distribution), while pink bars show the experimentally obtained numbers. The red curve is a Gaussian curve fitted to the experimental data. As can be clearly seen, the approach works well for up to 10 emitters per origami. At higher emitter numbers per cluster, the CoPS approach leads to an overestimation of this number.
n proof-of-concept studies, DNA origamis with varying emitter numbers (from 1 to 30) per origami were quantified. The plots below show the results obtained with DNA origami emitting in the red spectral range. The black bars indicate the expected distribution pattern of emitter number per origami (assuming a binomial distribution), while pink bars show the experimentally obtained numbers. The red curve is a Gaussian curve fitted to the experimental data. As can be clearly seen, the approach works well for up to 10 emitters per origami. At higher emitter numbers per cluster, the CoPS approach leads to an overestimation of this number.
Biological application example
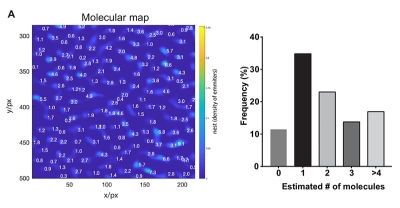 The γ-secretase complex is an intramembrane cleaving protease which cleaves more than a hundred transmembrane substrates. It`s aberrant activity is associated with Alzheimer`s disease and cancer. A detailed understanding of the complex`s structure and biology is required to target it with drugs.
The γ-secretase complex is an intramembrane cleaving protease which cleaves more than a hundred transmembrane substrates. It`s aberrant activity is associated with Alzheimer`s disease and cancer. A detailed understanding of the complex`s structure and biology is required to target it with drugs.
γ-Secretase is a tetrameric enzyme complex with 4 subunits (PSEN, NCT, APH1, PEN2), which are assembled in a 1💯1:1 ratio. While its structure is atomically resolved, knowledge about its distribution and dynamics in the plasma membrane so far comes mostly from biochemical data. For instance, it was unclear if γ-secretase exists as a monomer or if it forms higher-order multimers.
To answer this question, two complementary methods were employed in this publication to determine the γ-secretase stoichiometry at plasma membrane, namely counting of photobleaching steps and molecular counting by photon statistics (CoPS).
On the one hand, the photobleaching steps of GFP-PSEN1 were counted. On the other hand, the number of YFP-NCT was determined by CoPS. Additionally, the researchers validated that PSEN1 and NCT subunits are indeed present in a 1:1 stoichiometry. In summary, both complementary methods support the assumption that monomeric appearances are most abundant, followed by dimers of γ-secretase.
Publication: https://elifesciences.org/articles/56679
Downloads
Thank you for you inquiry.
We have received your message and will contact you as soon as possible.
×